Abstract
Background: Prognosis for children and adults with B-cell acute lymphoblastic leukemia (B-ALL), the most common pediatric malignancy, is heavily dependent on minimal residual disease (MRD) levels at the end of induction. Rates of detectable MRD are dependent on patient age and genomic subgroup. For patients with detectable MRD, the primary therapeutic adjustment is intensification of therapy if possible. Despite the incredible advances in MRD detection and clear associations with prognosis, uncovering the underlying biology of MRD could provide critical insight into mechanisms of therapy resistance and suggest research avenues for precise therapeutic approaches.
Methods: We collected samples from paired diagnostic and MRD time points from four patients with B-ALL, with MRD ranging from 0.15-0.84%. Based upon clinical MRD immunophenotype, we performed FACS sorting of lymphoblasts from each sample in order to isolate the rare MRD cells, and performed single-cell RNA sequencing (scRNA-seq) on all four pairs and single-cell ATAC sequencing (scATAC-seq) on two of these pairs. Data were compared across time points and integrated with publicly available datasets studying normal hematopoietic development. Raw scRNA-seq and scATAC-seq data were preprocessed using the Cell Ranger pipeline (10x Genomics, v.3.1.0). Seurat V4 (Hao et al., Cell 2021), Signac (Stuart et al., bioRxiv 2020), and MetaCell (Baran et al., Genome Biology 2019) were used to perform quality control (QC), normalize, identify highly variable features, perform dimension reduction, identify both discretized cell clusters and transient meta cells, and ultimately, visualize using uniform manifold approximation and projection (UMAP).
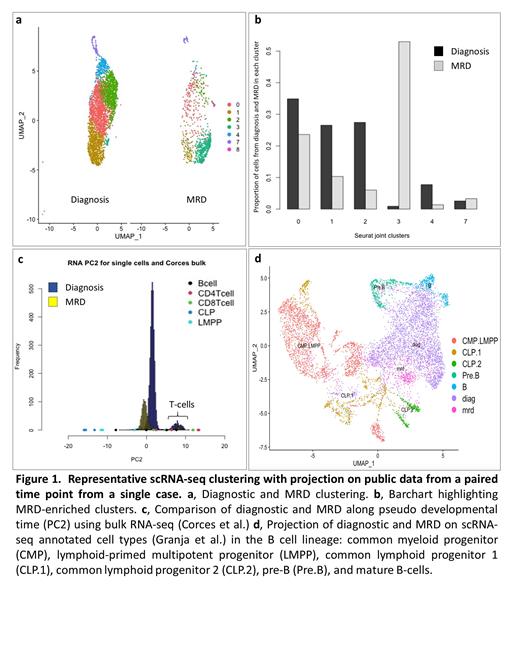
Results: Post scRNA-seq QC, the median number of cells per datasets was 2772, the median number of genes per dataset was 13855 and the median number of UMIs per cell was 6518. Post scATAC-seq QC, the median number of cells per datasets was 1157, the median number of peaks per dataset was 76190 and the median number of peaks per cell was 26642. We compared the diagnosis to MRD time points within each of the four cases. In each case, the transcriptionalprofile of the lymphoblasts changed from diagnosis to MRD timepoints (Fig. 1a), with specific transcriptional clusters enriched in the sorted MRD samples (Fig. 1b). To understand this difference, we compared the time points to publicly available datasets of normal hematopoietic development. First, we compared our data to published bulk RNA-seq data from sorted, well-defined hematopoietic development states (Corces et al., Nature Genetics 2016). We performed principal component analysis (PCA) on this data, and projected our generated scRNA-seq data onto the same principal component (PC) space. While the first PC captures the systematic artifact between the bulk and single-cell data, the second PC captures the developmental pseudo time. In each case, cells from sorted MRD show transcriptional profiles shifterd towards earlier progenitor cells on development pseudo time (Fig. 1c). We performed a similar analysis using scATAC-seq from a paired diagnosis and MRD sample, showing that the chromatin accessibility profile of MRD lymphoblasts reveals the same directional change towards patterns seen in earlier hematopoietic progenitor cells. We repeated a similar data integration on a published scRNA-seq dataset from well annotated normal hematopoietic developmental states (Granja et al. Nature Biotechnology 2019), which again revealed that the transcriptional patterns of MRD lymphoblasts align more closely with common lymphoid progenitors than pre-B cells. (Fig. 1d).
Conclusions: Using FACS sorting to isolated rare populations of MRD and subsequent single-cell analyses, we show that therapeutic pressure results in residual disease with an earlier progenitor transcriptional profile, likely determined by altered chromatin accessibility profile. Moving forward, we will explore the drivers of these changing transcriptomic and epigenomic profiles through expanding our number of cases to confirm initial findings, optimizing ATAC sequencing approaches to study the rare cell populations present in low-level MRD, and exploring specific transcription factor motifs driving these differences. Our findings have the potential to inform research into rational therapeutic approaches directed toward the specific biology of low level MRD in B-ALL.
No relevant conflicts of interest to declare.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal